Simulating Chemical Reaction Systems
The following python code simulates deterministic/stochastic mass action kinetics for a reaction system. The repositoriy can be found here
Importing the required libraries for the system
import numpy as np
from scipy.integrate import odeint
from scipy.special import comb,factorial
import matplotlib.pyplot as plt
Helping methods for the MassActionSystem class
- y : concentrations
- R : set of reactions
- k : corresponding rates
- t : time (redundant quantity required for odeint)
def odes(y,t,R,k):
l = R[:,0] # l, left complex
r = R[:,1] # r, right complex
return ((r-l).T).dot(arraypow(y,l.T)*k)
Method to compute :
For matrix and vector , the notation is the shorthand for the vector of monomials
def arraypow(x,A):
#Computes(\theta^A)
return np.prod(x**(A.T),axis=-1)
Reaction System class
Takes in the reactions in form of set of left, right complexes and corresponding rates. Takes in the species symbols, default species are prefixed by “S_”
class ReactionSystem(object):
"""docstring for ReactionSystem"""
def __init__(self, reactions, rates, species =None):
"""Each reaction in reactions is
a pair of complex (l,r) in l --> r"""
reactions = np.array(reactions)
rates = np.array(rates)*1.0
self.reactions = reactions
self.rates = rates
if species is None:
species =['S_'+str(i+1) for i in xrange(reactions.shape[2])]
self.species = species
def display_reactions(self):
"""
Puts together the set of reations in a string and returns it
"""
reaction_set =""
for i in xrange(self.reactions.shape[0]):
l = self.reactions[i][0]
r = self.reactions[i][1]
S = self.species
left_complex =""
right_complex=""
for j in xrange(len(S)):
if (l==0).all():
left_complex ="0 "
if (r==0).all():
right_complex ="0 "
if l[j]:
left_complex+= " " + str(l[j]) +"("+S[j]+")" + " +"
if r[j]:
right_complex+=" " + str(r[j]) +"("+S[j]+")" + " +"
reaction = left_complex[:-1] + " ------> " + right_complex[:-1] + " rate: "+ str(self.rates[i]) + "\n\n"
reaction_set +=reaction
return reaction_set[:-1]
Deterministic Mass Action System class
Inherits ReactionSystem and performs deterministic mass action kinetics on the system
class MassActionSystem(ReactionSystem):
"""docstring for MassActionSystem"""
def __init__(self, reactions, rates, species =None):
# super(MassActionSystem, self).__init__()
super(MassActionSystem,self).__init__(reactions, rates, species)
def set_concentrations(self,concentrations):
"""
Sets the concentration of the species
"""
concentrations = 1.0*np.array(concentrations)
if concentrations.shape[0] == self.reactions.shape[2]:
self.concentrations = concentrations
else:
print "Wrong Initialization: Shapes of concentrations and reactions dont match ",\
concentrations.shape[0], "!=", self.reactions.shape[1]
return self.concentrations
def dydt(self):
"""
Returns rates of concentrations change at the current concentration
"""
return odes(self.concentrations,0,self.reactions,self.rates)
def current_concentrations(self):
"""
Returns the concentrations after the latest run
"""
return self.concentrations
def run(self,t=1000,ts=100000,plot=False):
"""
Run it for time t with ts number of time steps
Outputs the concentration profile for the run
default values are 100000 and 1000000
"""
t_index = np.linspace(0, t, ts)
y = self.concentrations
output = odeint(odes, y, t_index, args= (self.reactions,self.rates))
self.concentrations = output[-1,:]
if plot:
for i in xrange(output.shape[1]):
label = self.species[i]
plt.plot(t_index, output[:, i], label=label)
plt.legend(loc='best')
plt.xlabel('t')
plt.title('Concentration Profile')
plt.grid()
plt.show()
return output
Stochastic Mass Action System
An implementation of the Gillespie Algorithm on a Reaction System. These slides might help understanding the stochastic simulation.
class StochasticSystem(ReactionSystem):
"""docstring for StochasticSystem
Gillespie Algorithm implementation
"""
def __init__(self, reactions, rates, species =None):
"""TODO: Maybe add a method to make sure reaction
coefficients are integers"""
super(StochasticSystem, self).__init__(reactions, rates, species)
def set_population(self,population):
population = 1.0*np.array(population)
if population.shape[0] == self.reactions.shape[2]:
self.population = population
else:
print "Wrong Initialization: Shapes of population and reactions dont match ",\
population.shape[0], "!=", self.reactions.shape[1]
return self.population
def current_population(self):
return self.population
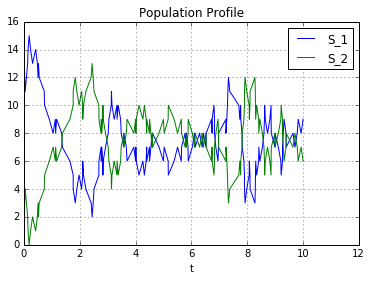
def run(self, t,seed=None,plot=False):
"""
Gillespie Implementation
"""
l = self.reactions[:,0]
r = self.reactions[:,1]
change=r-l
time =0
times =[]
output =[]
times.append(time)
output.append(self.population)
while time<t:
lam = np.prod(comb(self.population,l)*factorial(l),axis=-1)*self.rates
lam_sum = np.sum(lam)
dt = np.log(1.0/np.random.uniform(0,1))*(1.0/(lam_sum))
react = np.where(np.random.multinomial(1,lam/lam_sum)==1)[0][0]
self.population = self.population + change[react]
time += dt
output.append(self.population)
times.append(time)
output = np.array(output)
times = np.array(times)
if plot:
for i in xrange(output.shape[1]):
label = self.species[i]
plt.plot(times, output[:, i], label=label)
plt.legend(loc='best')
plt.xlabel('t')
plt.title('Population Profile')
plt.grid()
plt.show()
return times,output
Main
Simulating for simple examples:
def main():
reactions = [[[1,0,1],[0,2,0]], [[0,2,0],[1,0,1]]]
rates = [1,1]
system = MassActionSystem(reactions,rates)
print "Reactions:"
print system.display_reactions()
system.set_concentrations([-0.1 + 1/3.,0.2 + 1/3.,-0.1 + 1/3.])
print "Initial Concentrations:",system.current_concentrations()
system.run(t=10,ts=1000,plot=True)
print "Final Concentrations:",system.current_concentrations()
print "Final concentration change rates:", system.dydt(),"\n\n\n"
# Simulating Stochastic System
reactions = [[[1,0],[0,1]], [[0,1],[1,0]]]
rates = [1,1]
system = StochasticSystem(reactions,rates)
print "Reactions:"
print system.display_reactions()
population = [15,0]
system.set_population(population)
print "Initial Population:",system.current_population()
t,o = system.run(10,plot=True)
return
if __name__ == '__main__':
main()
Reactions:
1(S_1) + 1(S_3) ------> 2(S_2) rate: 1.0
2(S_2) ------> 1(S_1) + 1(S_3) rate: 1.0
Initial Concentrations: [ 0.23333333 0.53333333 0.23333333]
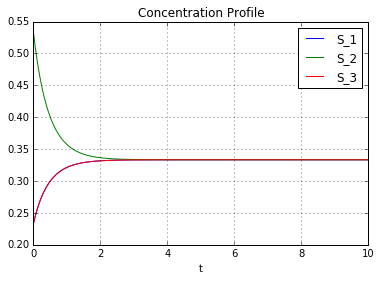
Final Concentrations: [ 0.33333333 0.33333333 0.33333333]
Final concentration change rates: [ 1.79787518e-10 -3.59575036e-10 1.79787518e-10]
Reactions:
1(S_1) ------> 1(S_2) rate: 1.0
1(S_2) ------> 1(S_1) rate: 1.0
Initial Population: [ 15. 0.]